Get High-Resolution ChIP-Exo-Seq Results for your Publication
Stop wasting precious samples and research time.
Our HPA-validated antibodies deliver superior specificity and reproducibility for chromatin immunoprecipitation applications.
97%
Specificity Rate
500+
Chip-Validated Antibodies
88%
Reduced Time-to-Results
1450+
ChIP Citations
ChIP Challenges: What’s Slowing You Down?
1. Non-Specific Binding
Generic antibodies often bind to off-target proteins, creating misleading signals and confounding your data interpretation. This leads to inconclusive or false results that can't be published.
2. Batch-to-Batch Variability
Inconsistent antibody performance between lots forces constant re-validation and protocol adjustments. Just when you optimize your workflow, the next batch fails.
3. Limited Sample Yield
Poor antibody efficiency means you need more starting material for successful ChIP experiments. This is particularly problematic with rare or patient-derived samples.
4. Unknown Epitope Accessibility
Many antibodies recognize epitopes that become inaccessible in crosslinked chromatin, resulting in weak or absent signals despite the target being present.
5. High Background Noise
Excessive background signal in ChIP-seq data creates analytical challenges and reduces peak detection confidence. This often requires complex bioinformatic correction.
6. Time-Consuming Optimization
Without validated ChIP antibodies, you might spend weeks or months adjusting conditions before getting usable data, delaying publications and discoveries.
The Atlas Antibodies ChIP-Exo-Seq Solution
Download the Application Note
ChIP-Exo-Seq by Atlas Antibodies
✅ Higher Resolution
Achieves near single-base precision for mapping binding sites.
✅ Reduced Noise
Exonuclease trimming eliminates background DNA, leading to cleaner data.
✅ Precise Localization
Provides more accurate mapping of transcription factor binding sites or other protein-DNA interactions
Establishing a Higher Benchmark for Antibodies Performance
Our antibodies demonstrate exceptional specificity and reproducibility in ChIP-Exo-Seq assays.
This ensures sharper, more accurate peak detection, marking exact protein-DNA binding sites.
Image: ChIP-Exo-Seq composite graph for Anti-ELF1 (HPA001755, Lot R00715) tested in K562 cells. Strand-specific reads (blue: forward, red: reverse) and IgG controls (black: forward, grey: reverse) are plotted against the distance from a composite set of reference binding sites. Plots are smoothed by a 21 bp moving average across 460 sites. The antibody exhibits robust target enrichment compared to a non-specific IgG control and precisely reveals its structural organization around the binding site. Data generated by Prof. B. F. Pugh´s Lab at Cornell University, Ithaca, NY, USA.
Ready to Elevate Your Chromatin Research?
Say goodbye to unreliable antibodies, inconsistent enrichment, and low-resolution data. Our ChIP-Exo–Seq-validated antibody collection delivers high specificity and reproducibility for precise mapping of protein-DNA interactions—saving your samples and your time.
Select from over 500 of expertly designed, ChIP-grade targets tailored for chromatin research.
🔬 Expert Support
Protocol optimization and technical guidance from ChIP specialists—our scientists at Atlas Antibodies and the Human Protein Atlas provide expert support and guidance to help you achieve reliable, reproducible results. From experiment design to troubleshooting, we're here to assist every step of the way.
FAQs: Your Questions Answered
-
What is ChIP-Exo-Seq, and how does it differ from ChIP-Seq?
ChIP-exo seq is a refinement of ChIP-seq that uses exonuclease digestion to improve the resolution of protein-DNA interaction mapping. The exonuclease trims DNA in a 5' to 3' direction, stopping at the protein-DNA crosslink. This results in single-base resolution of binding sites, compared to the broader peaks typical of ChIP-seq. It reduces background noise and provides more precise localization of binding sites.
-
What are the key advantages of ChIP-Exo-Seq over traditional ChIP-Seq?
- Single-base resolution: ChIP-exo seq provides near-base-pair precision for mapping binding sites.
- Reduced noise: Exonuclease digestion eliminates nonspecific DNA, leading to cleaner data.
- More accurate peak calling: Peaks are sharper and better defined compared to ChIP-seq.
- Higher reproducibility: The improved precision leads to better reproducibility across experiments.
-
What types of applications can benefit from ChIP-Exo-Seq?
ChIP-exo seq is particularly useful for:
- Identifying transcription factor binding sites with high precision.
- Mapping histone modifications or other epigenetic markers at single-base resolution.
- Understanding cis-regulatory elements in gene regulation.
- Enhancing functional studies in comparative genomics or between different cell states.
-
How does the data analysis for ChIP-Exo-Seq differ from ChIP-Seq?
In ChIP-exo seq, the data consists of sharp peaks marking the exonuclease stop points, requiring specialized analysis tools.
The analysis pipeline involves:
Mapping sequencing reads to the reference genome.
Calling precise peaks using algorithms that account for single-base resolution.
Comparing sharp peaks to motifs or regulatory elements for functional interpretation. Unlike ChIP-seq, the narrower peak width makes false positives less likely.
-
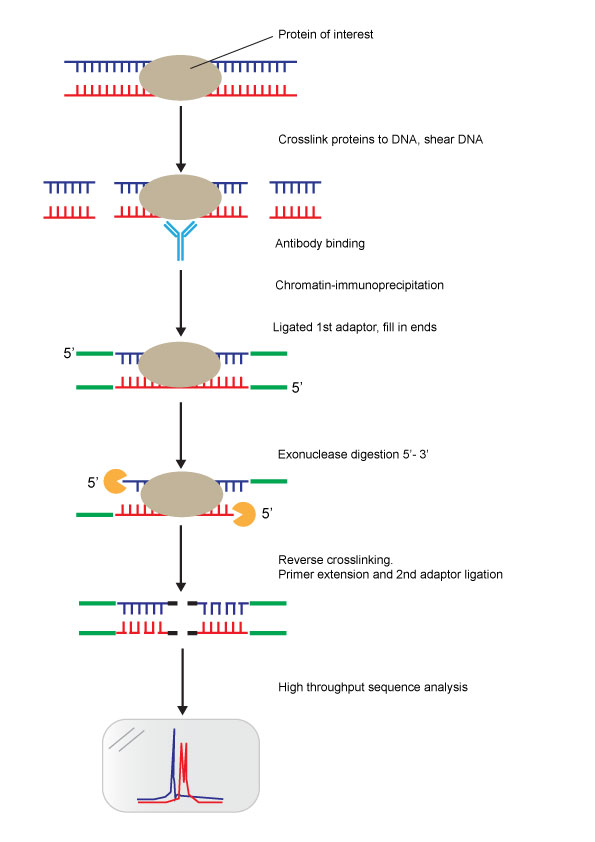
ChIP-Exo-Seq Protocol
Protocol Overview:
The method combines chromatin immunoprecipitation (ChIP) with next-generation sequencing to identify binding sites of DNA-associated proteins across the genome.
Steps in the Protocol:
Crosslink and shear DNA
Proteins of interest are crosslinked to DNA to preserve protein-DNA interactions.
The DNA is fragmented into smaller pieces using shearing methods (e.g., sonication).
Chromatin immunoprecipitation (ChIP)
Specific antibodies are used to bind the protein of interest, isolating the DNA-protein complex.
Immunoprecipitation is performed to pull down these complexes.
Adaptor ligation
A first adaptor is ligated to the ends of the sheared DNA fragments.
The ends are filled in to create blunt ends for downstream steps.
Exonuclease digestion (5' to 3')
An exonuclease enzyme digests the DNA in a 5' to 3' direction.
Digestion stops precisely at the point where the protein is crosslinked to DNA, leaving single-stranded DNA ends.
Reverse crosslinking and adaptor addition
The protein-DNA crosslinks are reversed, freeing the DNA.
The DNA is extended using primer extension, and a second adaptor is ligated to the opposite end.
High-throughput sequencing analysis
The processed DNA is subjected to high-throughput sequencing to map the protein-DNA binding sites.
The resulting sequencing data identifies sharp peaks, representing precise protein-DNA interaction sites at single-base resolution.