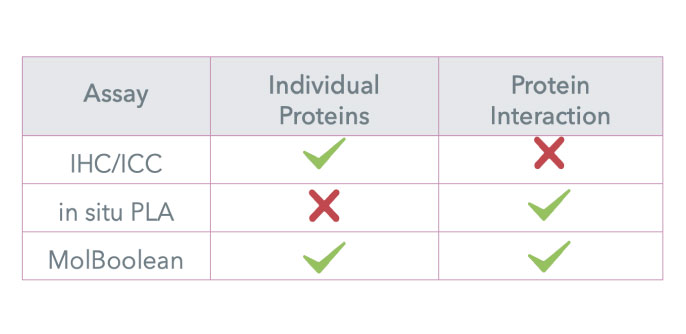
MolBoolean™ - Beyond Proximity Ligation

Comparative detection of EMD, LMNB1, and their complex in MCF7 cells using MolBoolean and in situ PLA.
Left panel: MolBoolean staining enables concurrent detection of free EMD, free LMNB1, and the EMD–LMNB1 proteins complex, providing multiplexed molecular interaction data within individual cells. This approach distinguishes between unbound and interacting protein pools.
Right panel: in situ Proximity Ligation Assay (inPLA) exclusively detects the EMD–LMNB1 complex, without resolving the presence of free, non-interacting EMD or LMNB1.
A universal kit that works with the customer’s choice of primary antibodies.
Designed for detection with one mouse and one rabbit primary antibody.
The kit is customizable for most species upon request, providing flexibility for diverse research applications.
Stop compromising. Co-IP loses location. Standard PLA creates ambiguity. FRET has limitations in fixed samples.
Listen to our Principal Scientist, Mikael Malmqvist PhD as he discusses the benefits of Atlas Antibodies' new technology for protein-protein interaction: MolBoolean.
MolBoolean™ analysis of the interaction (white) between ACE2 (magenta) and TMPRSS2 (green) in human kidney, using the monoclonal anti-ACE2 (Cat. AMAb91259) and the polyclonal anti-TMPRSS2 (Cat. HPA035787) antibodies from Atlas Antibodies AB.
Image shows the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 51% free ACE (magenta), 17% free TMPRSS2 (green) and 32% ACE/TMPRSS2 complex (white).
Data is normalized to total target protein levels (total RCPs).
MolBoolean™ analysis of the interaction (white) between E-cadherin (cat. AMAb90862, magenta) and β-catenin (cat. HPA029159, green) in MCF7 cells, showing the relative quantification of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels: 32% free E-cadherin (magenta), 19% free β-catenin (green), 49% E-cadherin/β-catenin complex (white).
Data is normalized to total target protein levels (total RCPs).
MolBoolean™ analysis of the interaction (white) between SATB2 (magenta) and HDAC1 (green) in human colon, using the monoclonal anti-SATB2 (Cat. AMAb90682) and the polyclonal anti-HDAC1 (Cat. HPA029693) antibodies from Atlas Antibodies AB.
Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
MolBoolean™ analysis of the interaction (white) between EMD (magenta) and LMNB1 (green) in MCF7 cells, using the monoclonal anti-EMD (Cat. AMAb90562) and the polyclonal anti- LMNB1 (Cat. HPA050524) antibodies from Atlas Antibodies AB.
Image shows the spatial location of free versus interacting protein fractions, indicated by the detection of rolling circle products (RCPs) in either one or two fluorescent channels.
Prof. Ola Söderberg, Uppsala University, Sweden (inventor of the MolBoolean technology) "Over the last two decades we have developed several methods to visualize proximity between proteins, MolBoolean being the latest and most advanced method. In contrast to PLA, it provides the ability to simultaneously monitor both the levels of free proteins and their interactions in fixed cells and tissue. The MolBoolean method was designed to incorporate targeting of both free and interacting proteins with one unified approach, instead of relying on orthogonal methods, keeping the efficiency in their reporting more similar."
Dr. Ilana Kotliar and Dr. Yuanhuang Chen, Prof. Thomas Sakmar, Laboratory at Rockefeller University, New York, USA "The MolBoolean fluorescence in situ protein proximity detection system exceeded our expectations as a robust analytical method to detect protein-protein interactions in cell membranes. With validated antibodies, the system could quantitate both individual components and complexes. Benchmarking and workflow were straightforward. We used chamber slides and the CellProfiler imaging and data processing pipelines. The results are spectacular."
Dr. Inés Rodríguez González and Prof. Dr. Ralph Kehlenbach, Georg-August-University, Göttingen, Germany "MolBoolean analysis is a very interesting approach to obtain quantitative data of in situ protein interactions that are probably very weak or transient and difficult to capture by conventional biochemical methods. The biggest advantage of MolBoolean is that, in addition to detecting two proteins in close proximity, it allows the quantification of the individual pools of protein 1 and 2, facilitating a direct comparison with the interacting fraction. The MolBoolean protocol is also informative and easy to follow. Deconvolution of microscopy images to further increase their resolution and the analysis of the obtained RCPs can be performed in open access software such as FIJI and Cell Profiler. All in all, we believe that MolBoolean analysis is a technology with many advantages and has the potential to be exploited and applied in different areas of cell biology.”
Prof. Rafael Franco, Network Center for Biomedical Research in Neurodegenerative Diseases, University of Barcelona, Spain. "Since the hypothesis of GPCR heteromers at the cell surface there has been much advance, except in what concerns the relative numbers of interacting versus non-interacting receptors. We tested MolBoolean and we are surely happy with the experience, and the results. We used the kit to study the expression levels of A2A (A2AR)/dopamine D2 (D2R) receptor heterodimers in the striatum of rat and non-human primate models of Parkinson’s disease. MolBoolean allowed us to quantify the levels of receptor heterodimers. More importantly, the ability to also detect non-interacting receptors enabled us to determine the percentage of total receptors that are forming these heterodimers, shedding more light on how A2AR mediate their antagonistic regulation on motor control in the brain. MolBoolean workflow and protocol were straightforward and easy to follow.
How MolBoolean differs from PLA?
MolBoolean™ is a kit for in situ protein proximity analysis in tissue and cells developed by Atlas Antibodies. Unlike traditional methods, it provides spatial quantitative analysis of protein-protein interactions by the simultaneous detection of free and interacting proteins (~40 nm proximity).
The “Molecular Boolean" Logic
The MolBoolean™ assay applies a Boolean logic at a molecular level to distinguish between free and interacting proteins, mapping protein interactions with high spatial specificity.
The OR logic identifies either protein A or B independently, signaling that they are present but not necessarily interacting.
The AND logic is applied when proteins A and B are interacting; they are detected together as complex AB, indicating a combined "true" interaction.
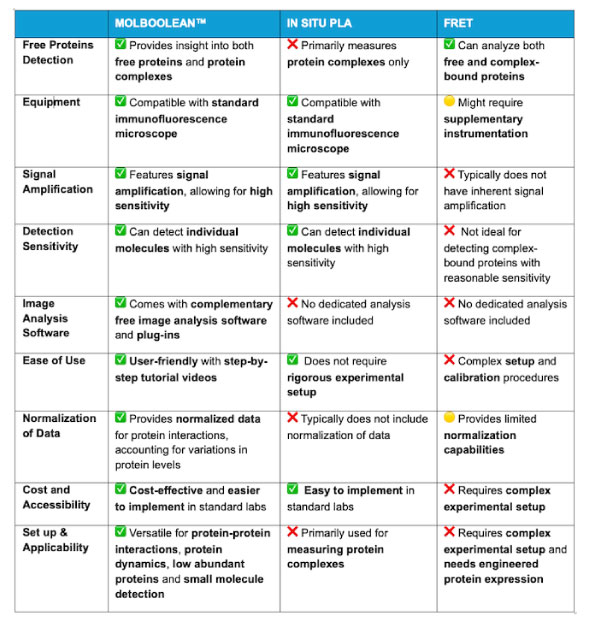
What are the benefits of using MolBoolean™ compared to traditional methods for studying protein-protein interactions?
Comparative table that highlights the advantages of MolBoolean™ over in situ PLA and FRET for studying protein-protein dynamics:
Why choose MolBoolean?
MolBoolean provides spatial quantitative analysis of protein-protein interactions by the simultaneous detection of free
and interacting proteins. MolBoolean™ offers significant benefits in fields requiring complex spatial interaction and quantification data such as:
Product Description
MolBoolean™ Mouse/Rabbit is a kit for in situ protein proximity analysis in tissue and cells. The MolBoolean™ assay utilizes a proprietary oligonucleotide setup that enables the simultaneous detection of both free and interacting (~40 nm proximity) fractions for two proteins of interest (protein A, protein B and interaction proteins AB). It uses two different immunofluorescent detection reporters with maximum emission wavelengths at 590 nm (ATTO565, TxRed filter or similar) and at 664 nm (ATTO647N, Y5 filter or similar), respectively.
MolBoolean™ Mouse/Rabbit can be performed with the user’s primary antibodies of choice (raised in mouse and rabbit). MolBoolean™ relies on anti-mouse and anti-rabbit secondary proximity probes and rolling circle amplification (RCA) as a mean to amplify signal.
A series of molecular steps, performed in the MolBoolean™ assay, incorporates information into the amplified products indicating antibody target engagement of individual versus interacting proteins. This information is converted to fluorescent signals by the binding of detection reporters, allowing visualization on a conventional fluorescent microscope. An image analysis software is provided to segment and differentiate fluorescent signals, enabling the relative quantification of free versus interacting fractions for the two analyzed protein targets in the sample.
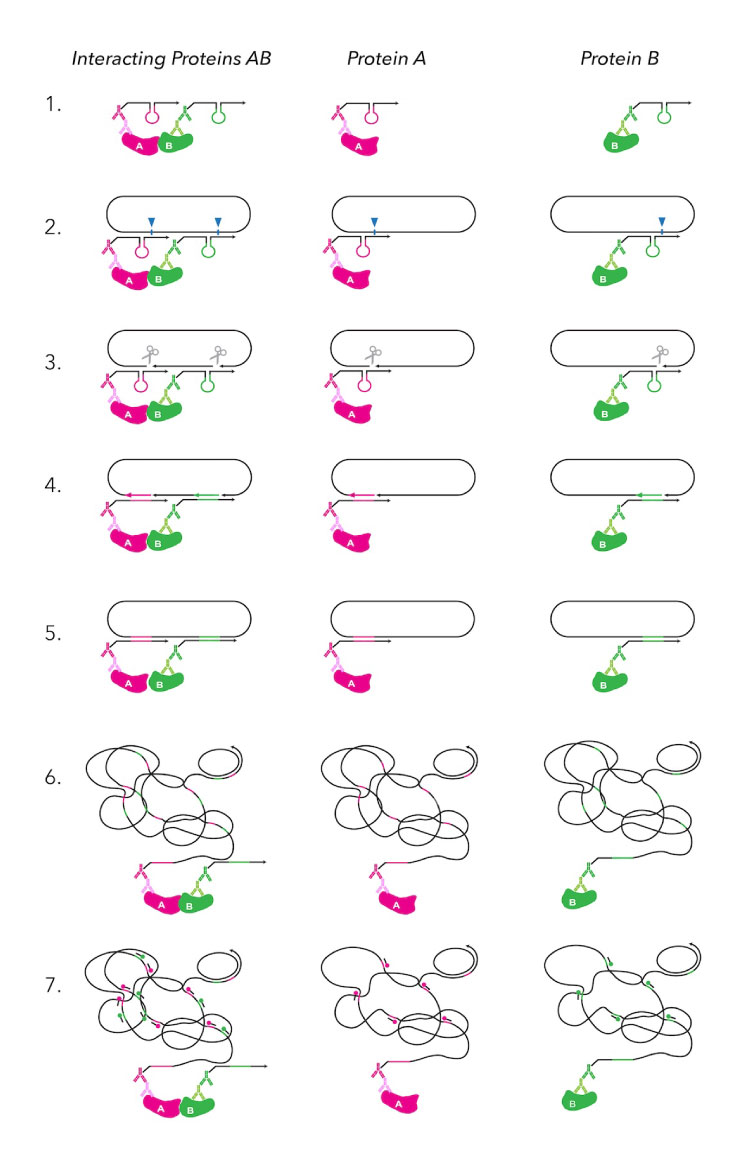
MolBoolean Assay Steps and Workflow
The MolBoolean™ assay follows a 2-days 7-steps workflow to determine if target proteins are interacting (AB) or present individually (A, B).
Day 1 (2 hours): blocking and primary antibodies incubation.
Day 2 (7-8 hours): probe binding, tagging, amplification, and detection.
(STEP 1) Proximity probes bind to primary antibodies attached to the proteins.
(STEP 2) Proximity probes link to the DNA circle.
(STEP 3) DNA nicking prepares the DNA for tagging.
(STEP 4) Tag oligos are added to mark the proteins.
(STEP 5) DNA ligation connects the DNA segments.
(STEP 6) Rolling circle amplification (RCA) boosts the signal for easier detection.
(STEP 7) Detection reveals both interacting and separate proteins.
What's Included in the MolBoolean Kit?
The MolBoolean™ kit includes 15 separate tubes: enzymes (nickase, ligase, polymerase), buffers, oligos for tagging and detection, secondary antibody probes and additional reagents such as blocker, and diluent.
1:Blocker (4x) - 2:Diluent (1x) - 3:Probe A (80x) - 4:Probe B (80x) - 5:Circle (40x) - 6:Additive (40x) - 7:Buffer A (10x) - 8:Nickase enzyme (80x) - 9:Buffer B (8x) - 10:Tag oligos (40x) - 11:Ligase enzyme (80x) - 12:Polymerase enzyme (40x) - 13:Buffer C (5x) - 14:Detection oligos (40x) - 15:Buffer D (10x).
The total volume of 4.8 ml, is sufficient for approx. 120 assays in cells (40 μl/assay) and 60 assays in tissue (80 μl/assay).
Can MolBoolean be used with any primary antibodies?
MolBoolean is a kit that can be used with the customer’s choice of primary antibodies (mouse /rabbit). MolBoolean is compatible with protocols already established in the lab for primary antibodies. This flexibility allows researchers to seamlessly integrate MolBoolean into their existing workflows without requiring the purchase of specialized antibodies, making it a convenient and adaptable tool for studying protein interactions and free protein levels.
Be aware that the specificity of the primary antibodies can affect the accuracy of MolBoolean results. It is important to validate the primary antibodies to ensure that they specifically target the proteins of interest before applying the MolBoolean technology. We recommend the use of Enhanced validated primary antibodies.
📖 Peer Reviewed Publications
Rivas-Santisteban R, et al, (2024) GPR88 impairs the signaling of kappa opioid receptors in a heterologous system and in primary striatal neurons. Neuropharmacology. 2024 Nov 27:110242. doi: 10.1016/j.neuropharm.2024.110242. Epub ahead of print. PMID: 39613254.
Kotliar IB, et al, (2024) Multiplexed mapping of the interactome of GPCRs with receptor activity-modifying proteins. Sci Adv. 2024 Aug 2;10(31):eado9959. Epub 2024 Jul 31. PMID: 39083597; PMCID: PMC11290489.
Malmqvist M, et al, (2024) Quantifying dopamine D2 and adenosine A2A receptor interactions in rat brain using MolBoolean™ technology. #8217 Society for Neuroscience, SfN, (2024).
Raykova D, et al, (2023) A method for Boolean analysis of protein interactions at a molecular level. Nat Commun. 2023 Sep 6;14(1):5450. doi: 10.1038/s41467-023-41325-3. Erratum for: Nat Commun. 2022 Aug 13;13(1):4755. doi: 10.1038/s41467-022-32395-w. PMID: 37673885; PMCID: PMC10482831.
Rivas-Santisteban R, et al, (2023) Boolean analysis shows a high proportion of dopamine D2 receptors interacting with adenosine A2A receptors in striatal medium spiny neurons of mouse and non-human primate models of Parkinson’s disease. Neurobiol Dis. 2023 Nov;188:106341.doi: 10.1016/j.nbd.2023.106341. Epub 2023 Oct 31. PMID: 37918757.