Orthogonal Validation in IHC
One of the key factors for reproducible research is to use antibodies that have been thoroughly validated for the application and model system that will be used. We are proud to be the first company to offer our customers enhanced antibody validation in IHC on a broad scale.
Our antibodies are developed by the Human Protein Atlas and are rigorously evaluated for specificity and performance and are characterized in several applications. The antibodies are validated for IHC in all major tissues and organs as well as the 20 most common cancer types. An additional layer of validation, enhanced validation, is performed following the Human Protein Atlas interpretation of the recommendations by the International Working Group for Antibody Validation (IWGAV) published in Nature Methods by Uhlén et al.
Unique orthogonal validation in IHC
Orthogonal validation is an enhanced validation method where the antibody staining is verified by a non-antibody based method. At Atlas Antibodies, this is applied in IHC by analyzing the relative expression of the target protein in different samples using IHC and transcriptomics. Here, the antibody staining is compared to RNA-Seq data for the same samples, using both positive and negative or low expression samples. The specificity of the antibody is verified when the antibody signal in IHC correlates to the RNA levels in the evaluated samples.
Benefits of orthogonal validation in IHC comparing protein expression and transcriptomics
Verifying antibody binding by comparing the relative protein expression in IHC to transcriptomics has several advantages:
- The antibody binding can be evaluated without the need for knocking out or down the gene, which may be difficult or unfeasible in IHC depending on the protein and species that are studied.
- This method is useful also for validating antibodies targeting unknown or not well-studied proteins, where little or no literature is available.
- A useful approach for proteins that are essential for the cell and not possible use downregulation the target gene.
- In contrast to downregulation, testing in high and low expression samples also gives an indication of potential cross-reactivity.
Interpreting the validation data
In the data presented for the enhanced validation, the IHC stainings from two samples are displayed side by side for comparison. Below the staining images, bar charts illustrate the TPM values from RNA-seq analysis in the same tissues. The expected result is a correlation of the intensity level of the staining in IHC to the RNA levels in the same tissues.
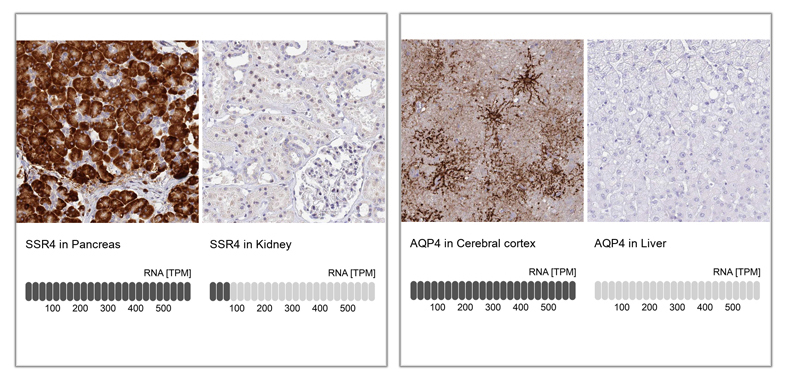 Examples of enhanced validation in IHC, comparing antibody signal to RNA-Seq data, using tissues with high and low expression of the corresponding protein. A: IHC analysis in human cerebral cortex and liver tissues using Anti-AQP4 antibody (HPA014784). Below the two staining images, bar charts depicting the TPM values for AQP4 is presented. B: IHC analysis in human pancreas and kidney tissues using Anti-SSR4 antibody (HPA045209). Below the staining images, bar charts Illustrates the level of TPM values for SSR4 from RNA-seq analysis in Pancreas and Kidney. In both cases, there is strong staining in the tissues with high RNA expression of the protein and low or absence of staining in tissues with low or no expression of the protein.
Examples of enhanced validation in IHC, comparing antibody signal to RNA-Seq data, using tissues with high and low expression of the corresponding protein. A: IHC analysis in human cerebral cortex and liver tissues using Anti-AQP4 antibody (HPA014784). Below the two staining images, bar charts depicting the TPM values for AQP4 is presented. B: IHC analysis in human pancreas and kidney tissues using Anti-SSR4 antibody (HPA045209). Below the staining images, bar charts Illustrates the level of TPM values for SSR4 from RNA-seq analysis in Pancreas and Kidney. In both cases, there is strong staining in the tissues with high RNA expression of the protein and low or absence of staining in tissues with low or no expression of the protein.
Method
The orthogonal validation method applied in IHC at Atlas Antibodies is performed as follows:
- RNA-Seq data for up to 37 normal tissues is obtained from the Human Protein Atlas database, to assess where the target gene is expressed.
- Two tissues are chosen for the validation, one with high RNA expression and the other with low or no RNA expression of the target. The tissues are chosen so that there is at least a five-fold difference between the RNA expression in the high and low samples.
- IHC staining were performed in the two tissues at the same time using the same dilutions with our standard IHC protocol.
- The correlation between antibody staining intensity and corresponding mRNA levels are manually evaluated and compared.
- Antibody performance is validated when the antibody signal in IHC correlates with RNA levels in both high and low expression samples.
Important considerations
- When determining what constitutes high and low expression, keep in mind that this is relative and that there has to be a significant difference between the samples. At Atlas Antibodies we have used a five-fold difference as criteria.
- Comparing the difference in expression levels is relevant between different tissues for a specific target, not comparing the difference in expression levels between different targets.
- Antibodies targeting housekeeping proteins may be difficult to validate using this orthogonal method, as they may show similar expression in all or most tissues.
- When studying secreted proteins, it is worth noting that the RNA levels and protein expression may not correlate. RNA levels in a specific tissue may be high, but as the protein is secreted, it can be detected at the protein level in another location.
