The Five Methods of Enhanced Validation
 Genetic Validation
Genetic Validation
In this enhanced validation method, antibody specificity is confirmed by downregulating the target protein on a genetic level using siRNA or CRISPR-Cas9.
Antibody specificity is confirmed when knockout or knockdown of the corresponding gene correlates with the absence or decrease of the antibody signal.
Genetic validation using siRNA knockdown is a method used at Atlas Antibodies to validate antibodies in Western blot. The method can also be applied for antibody validation in ICC-IF.
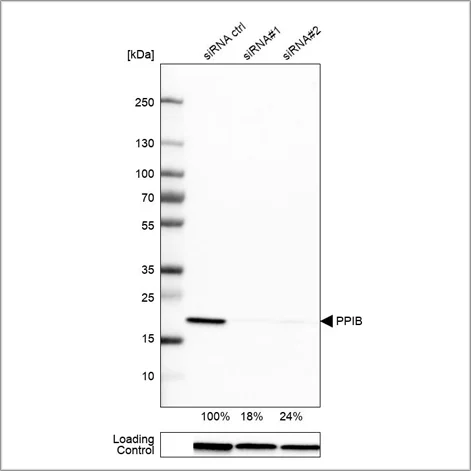
Example of genetic validation by siRNA knockdown in Western blot using the Anti-PPIB antibody (HPA012720). U-251 cells have been transfected with control siRNA and two target-specific siRNA probes. Downregulation of antibody signal confirms target specificity. The remaining intensity relative control lane is indicated as a percentage.
 Orthogonal Validation
Orthogonal Validation
In this enhanced validation method, the antibody is validated by comparing the results with a non-antibody-based method across multiple samples.
One orthogonal approach is to compare antibody staining intensities to RNA-Seq data from the same samples, over multiple tissues or cells with varying expression of the target protein. Antibody specificity is confirmed when the antibody signal matches RNA levels in the evaluated samples.
For each antibody, two tissues or endogenous cell lines are chosen for the validation, one with high RNA expression and the other with low or no RNA expression of the target.
Enhanced validation by comparing the antibody signal to RNA-Seq data is used at Atlas Antibodies to validate antibodies in WB and IHC. The method can also be applied to ICC-IF.
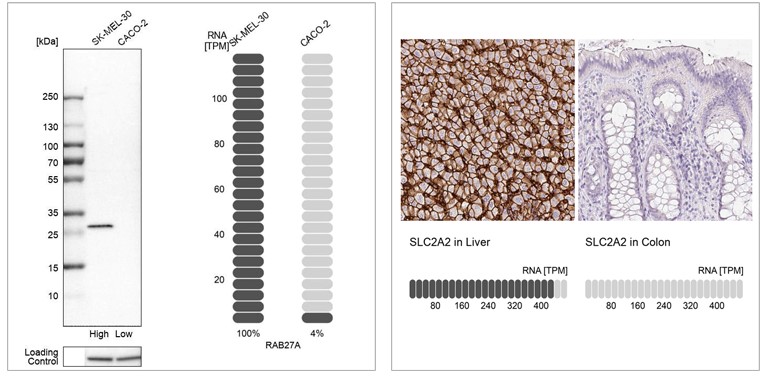 Examples of orthogonal validation in WB and IHC. Left: WB analysis in human cell lines SK-MEL-30 and Caco-2 using Anti-RAB27A antibody (HPA001333). Corresponding RAB27A RNA-seq data (TPM values) are presented for the same cell lines. Right: IHC staining of liver and colon tissues using the Anti-SLC2A2 antibody (HPA028997). The corresponding RNA-Seq data (TPM values) for the same tissues are presented below. In both examples, samples with known high and low RNA expression are chosen.
Examples of orthogonal validation in WB and IHC. Left: WB analysis in human cell lines SK-MEL-30 and Caco-2 using Anti-RAB27A antibody (HPA001333). Corresponding RAB27A RNA-seq data (TPM values) are presented for the same cell lines. Right: IHC staining of liver and colon tissues using the Anti-SLC2A2 antibody (HPA028997). The corresponding RNA-Seq data (TPM values) for the same tissues are presented below. In both examples, samples with known high and low RNA expression are chosen.
Orthogonal Validation in IHC
Dig deeper - learn how our unique orthogonal validation is applied in IHC.
Learn more
 Validation by Independent Antibodies
Validation by Independent Antibodies
In this enhanced validation method, antibody specificity is demonstrated by comparing two antibodies targeting different regions of the same protein.
If the two antibodies generate a similar staining pattern when compared in a set of relevant tissues, the antibodies validate each other.
Independent antibody validation can be applied in IHC, WB, and ICC-IF setups. The approach requires:
- Exact knowledge of the antigen sequence for each antibody.
- At least two antibodies with non-overlapping antigen sequences for each protein target.
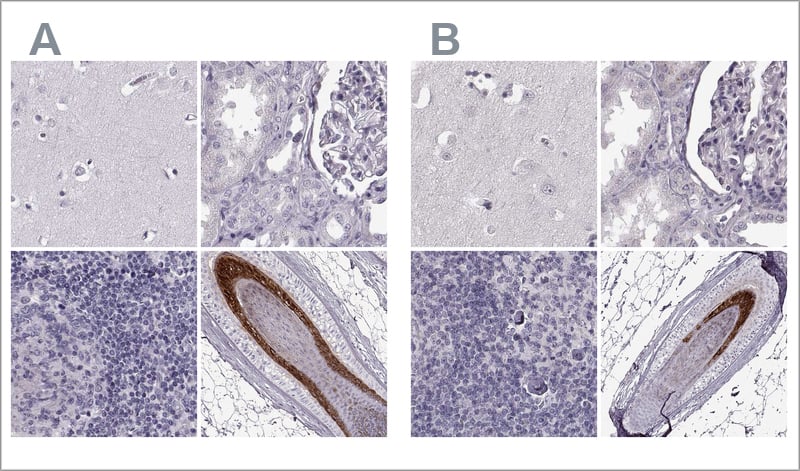
Examples of independent antibody validation in IHC. The two Anti-TCHHL1 antibodies HPA063483 (left, A) and HPA042579 (right, B) target different regions of TCHHL1. Antibody stainings across relevant positive and negative tissues are similar between the two, and the antibodies validate each other's staining pattern in IHC. Here, you can see IHC stainings of each of the two antibodies of the cerebral cortex, kidney, lymph node, and skin.
 Recombinant Expression Validation
Recombinant Expression Validation
In this enhanced validation method, the antibody binding is verified using an over-expressed or tagged version of the target protein.
When over-expressing the target protein in a cell line, the antibody is validated by comparing the signal from the over-expressed version with the unmodified endogenous target protein. This approach can be applied in WB.
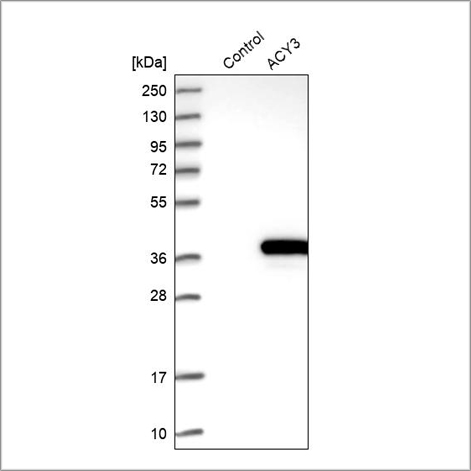
Example of recombinant expression validation in Western blot using the Anti-ACY3 antibody (HPA039219). Lane 1: marker, lane 2: negative control (vector only transfected HEK293T lysate), lane 3: ACY3 over-expression lysate (Co-expressed with a C-terminal myc-DDK tag (~3.1 kDa) in mammalian HEK293T cells, LY408962).
Alternatively, the target protein is tagged by an affinity tag or a fluorescent protein. The pattern displayed by the tagged target protein is matched to the antibody signal. A match confirms that the antibody is recognizing its target protein. Validation by tagged proteins can be applied in ICC-IF.
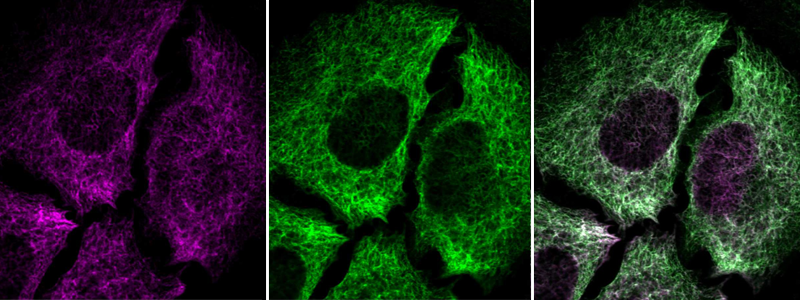
Immunofluorescent staining of transgenic HeLa cells using Anti-NES antibody (HPA006286) shows positivity in intermediate filaments (in green). Antibody staining overlaps with GFP-tagged nestin protein (in purple).
 Migration Capture MS Validation
Migration Capture MS Validation
In this enhanced validation method, the staining pattern and the protein size detected by the antibody is compared with results obtained by a capture Mass Spectrometry (MS) method.
Antibody specificity is confirmed when the size detected by the antibody is equivalent to the size of the corresponding target protein detected in migration capture MS.
Validation by Migration Capture MS will be introduced for selected antibodies.





 Examples of orthogonal validation in WB and IHC. Left: WB analysis in human cell lines SK-MEL-30 and Caco-2 using Anti-RAB27A antibody (HPA001333). Corresponding RAB27A RNA-seq data (TPM values) are presented for the same cell lines. Right: IHC staining of liver and colon tissues using the Anti-SLC2A2 antibody (HPA028997). The corresponding RNA-Seq data (TPM values) for the same tissues are presented below. In both examples, samples with known high and low RNA expression are chosen.
Examples of orthogonal validation in WB and IHC. Left: WB analysis in human cell lines SK-MEL-30 and Caco-2 using Anti-RAB27A antibody (HPA001333). Corresponding RAB27A RNA-seq data (TPM values) are presented for the same cell lines. Right: IHC staining of liver and colon tissues using the Anti-SLC2A2 antibody (HPA028997). The corresponding RNA-Seq data (TPM values) for the same tissues are presented below. In both examples, samples with known high and low RNA expression are chosen.




